Over the years I have looked
closely at the peopling of the Americas on this blog. In particular, I have
focused on examining pre-Clovis archaeological sites to see how the evidence
stacks up. For instance, I have looked at the following sites:
Monte Verde in Chile (see here)
Arroyo del Vizcaíno, Uruguay (see here)
Meadowcroft Rockshelter, Pennsylvania USA (see here)
Buttermilk Creek, Texas USA (see here)
Nugget Gulch, Yukon Canada (see here)
Blue Fish Caves, Canada (see here)
Santa Elina Rock Shelter, Mato Grosso, Brazil (see here)
Cerutti Mastodon site, California USA (see here)
One thing I have not done
however, is look at the genetic evidence for the peopling of the Americas in
any detail.
To do so I needed to understand
the science, underlying the academic DNA papers more thoroughly. I have
therefore, had to go back to school!
To analyse what a particular
genetics paper means in the wider context of the peopling of the Americas, I
needed to know what data palaeogeneticists collected and what it meant at a
basic level. I have now completed a basic study of the topic. As I went along I
took notes for my own reference. I thought these may be of interest to others
looking to read this type of academic paper. I have therefore put this post up
here in the hopes, that it may be, of some help, to others in a similar
situation to myself.
The basics
Our bodies are made of cells.
These are small building blocks that join, together, to make organs and
organ-systems or float freely in our circulatory system, for example blood
cells.
The function of each cell is
determined by a set of instructions contained in the nucleus of each cell. This
set of instructions is written in a chemical molecule called DNA.
DNA, or deoxyribonucleic acid, is
the hereditary material in humans and almost all other organisms. Nearly every
cell in a person’s body has the same DNA. Most DNA, is located in the cell
nucleus (where it is called nuclear DNA), but a small amount of DNA can also be
found in the mitochondria (where it is called mitochondrial DNA or mtDNA).
Mitochondria are structures within cells that convert the energy from food into
a form that cells can use.
The information in DNA is stored
as a code made up of four chemical bases: adenine (A), guanine (G), cytosine
(C), and thymine (T). Human DNA consists of about 3 billion bases, and more
than 99 percent of those bases are the same in all people. The order, or
sequence, of these bases determines the information available for building and
maintaining an organism, similar to the way in which letters of the alphabet
appear in a certain order to form words and sentences.
DNA bases pair up with each other, A with T and
C with G, to form units called base pairs. Each base is also attached to a
sugar molecule and a phosphate molecule. Together, a base, sugar, and phosphate
are called a nucleotide. Nucleotides are arranged in two long strands that form
a spiral called a double helix. The structure of the double helix is somewhat
like a ladder, with the base pairs forming the ladder’s rungs and the sugar and
phosphate molecules forming the vertical sidepieces of the ladder.
Stretch of DNA showing base
pairs.
In humans, the DNA is packed into 23 pairs of
homologous molecules called Chromosomes, for a total of 46 Chromosomes. Each of
the homologous chromosomes in a pair
is inherited from a different parent. So, we get half of our genetic material –
DNA - from each parent.
One set of human chromosomes (picture credit: US Library of
Medicine)
How do humans
differ from one another?
A gene is commonly defined as a
DNA sequence, on a particular chromosome, that has a function, meaning a class
of similar DNA sequences all involved in the same, particular, molecular,
function.
Alleles can defined as
“alternative forms” of a gene that can occur at the same locus, or place, in
the genome. Many Alleles can be caused by Single Nucleotide Polymorphisms, or
other changes such as deletions, transversion, or insertions.
A single-nucleotide polymorphism,
abbreviated to SNP is a variation in a single nucleotide that occurs at a
specific position in the genome.
For example, at a specific base
position in the human genome, the C nucleotide may appear in most individuals,
but in a minority of individuals, the position is occupied by an A. This means
that there is a SNP at this specific position, and the two possible nucleotide
variations – C or A – are said to be alleles for this position.
By any definition a gene must
involve more than one nucleotide base pair. Single nucleotide polymorphisms
(SNPs) thus do not occur at loci, but rather in and around loci.
SNP markers do not, therefore
occur “at” loci. SNP markers do have certain, alleles at set locations: that is
“sites” within the region of a locus.
If in a population only one
allele occurs at a site or locus, we say that it is monomorphic, or
monoallelic, in that population. If two alleles occur, as is common for SNPs,
we use the term diallelic (also known as biallelic). If many
alleles occur, the polymorphism is called polyallelic or multiallelic. When
there are just two alleles at a locus, the one with the smaller population
frequency is called the minor allele. In genetics, the term allele
“frequency”- which is strictly speaking a count - is used to mean relative
frequency, i.e. the proportion of all such alleles at that locus among the
members of a population; thus the term minor allele frequency is often used for
diallelic markers.
A polymorphic locus was originally defined as a
locus at which the least common allele occurs with a “frequency” of at least 1%
but a more appropriate definition would be a locus at which the most common
allele occurs with a “frequency” of at most 99%. Different alleles arise at a
locus as a result of mutation, or sudden change in the genetic material. Mutation is a
relatively rare event, caused for example by an error in replication or the
action of a mutagen. Thus all alleles are by origin mutant alleles, and a
genetic polymorphism was conceived of as a locus at which the frequency of the
least common allele has a frequency too large to be maintained in the
population solely by recurrent mutation. However, what is important at a locus
is the degree of polymorphism, and a locus in which there are 1,000
equifrequent alleles would be considered much more polymorphic than a locus at
which there are two alleles with frequencies 0.01 and 0.99. Many authors now
use the term mutation for any rare allele, and the term polymorphism for any
common allele.
A haplotype is the multilocus
analogue of an allele at a single locus. It consists of one allele from each of
multiple loci that are transmitted together from a parent to an offspring. So haplotypes are made up of multiple alleles (one from each locus). It is usual
nowadays to restrict the word haplotype to the case where all the loci involved
are on the same chromosome pair, so that all the alleles involved are on the
same chromosome.
If the alleles at one locus are
not distributed in the population independently of the alleles at another
locus, the two loci exhibit allelic association. If this association is a
result of a mixture of subpopulations (such as ethnicities or religious groups)
within each of which there is random mating, the association is often denoted
as “spurious”. In such a case there is true association, but the cause is not
of primary genetic interest. If the association is not due to this kind of
population structure, it is either due to linkage disequilibrium (LD) or
gametic phase disequilibrium (GPD); in the former case the loci are linked,
i.e. they co-segregate in families, in the latter case they are not linked,
i.e. they segregate independently in families.
Identity
The concept of allelic identity
is an important one. Alleles are identical by descent (IBD) if they are copies
of the same ancestral allele, and must be differentiated from alleles that are
physically identical but not (at least within the previous dozen or so
generations) ancestrally identical. Such alleles, when not IBD, are identical
in state (IIS) or more commonly, nowadays identical by state (IBS). Here these
alleles, are ancestrally, but not physically, different.
Mitochondrial DNA
Although most DNA is packaged in
chromosomes within the nucleus, mitochondria also have a small amount of their
own DNA. This genetic material is known as mitochondrial DNA or mtDNA.
Mitochondrial DNA is inherited, unchanged directly from the female parent.
Mitochondria, are cellular
organelles within eukaryotic cells that convert chemical energy from food into
a form that cells can use, adenosine triphosphate (ATP). These organelles are
found in most cell types, including bone.
As, a large number of,
mitochondria and hence, mitochondrial DNA is found in most cells, there is a
relatively large amount in most samples from living or deceased individuals,
available for study, once extracted.
Diagrammatic representation of the position of mtDNA in cells from Wikipedia commons (2019)
Since human mtDNA evolves faster
than nuclear genetic markers, it has become a mainstay of phylogenetics and
evolutionary biology. The fact that mitochondrial DNA is maternally inherited
enables genealogical researchers to trace maternal lineage far back in time.
By looking at the SNPs in mtDNA,
haplogroups and haplotypes can be determined. The order and number of the SNP
changes allows geneticists to construct a phylogenetic tree showing the
relatedness in both time and space of the various haplogroups and haplotypes.
It has therefore, permitted an
examination of the relatedness of populations, and so has become important in
anthropology and biogeography.
y-DNA
Only males have a Y-chromosome, thus
making their 23rd chromosome pair XY, whereas women have two X chromosomes
in their 23rd pair. The Y-chromosome is almost 60 million base pairs
long and there is only one per cell. A man's patrilineal ancestry, or male-line
ancestry, can be traced using the DNA on his Y chromosome (Y-DNA), because the
Y-chromosome is transmitted father to son nearly unchanged.
Single nucleotide polymorphisms
(SNPs)
As was the case in mtDNA,
single-nucleotide polymorphisms (SNPs) also occur in y-DNA. These single
changes to a nucleotide in a DNA sequence will, when taken together confirm
haplogroup and haplotype.
Typical, commercial, y-DNA SNP
tests test about 20,000 to 35,000 SNPs, while academic researchers use far more
e.g. Fu et al. (2016) used between 200,000 and ca. 800,000 SNPs for the
delineation of haplotypes and relationships between ancient individuals.
Again, as in mtDNA, haplogroups
and haplotypes can be determined, and the order and number of the SNP changes
allows geneticists to construct a phylogenetic tree showing the relatedness in
both time and space of y-DNA haplogroups and haplotypes. Different branches of
this tree are different haplogroups. Most haplogroups can be further subdivided
multiple times into sub-clades and finally haplotypes.
Once more, this type of DNA also,
permits an examination of the relatedness of populations, and so has become
important in anthropology and biogeography.
For example, commercial DNA
analysis has brought up some interesting results as noted by Bettinger (2016): “All
human men descend in the paternal line from a single man dubbed Y-chromosomal
Adam, who lived probably between 200,000 and 400,000 years ago. ..Most significant of these new
discoveries was in 2013 when the haplogroup
A00 was discovered, which required theories
about Y-chromosomal Adam to be significantly revised.”
If we compare the ease with which
y-DNA and mtDNA can be collected some important facts emerge. For the y-DNA,
there is only one copy per cell, in the nucleus. If you recall mtDNA resides in
the mitochondria of cells. On average, there are 2000 mitochondria per cell.
Therefore, it relatively, easy to find undamaged mtDNA, in even ancient samples.
Conversely DNA analysis carried
out to examine the y-DNA looks at the diagnostic regions of the 60 million base
pairs of the y-chromosome to determine haplogroup and type. To do so, depends
on extracting enough DNA from these regions within those 60 million base pairs
for analysis. For highly degraded remains, it's highly unlikely that enough of
the right Y survives for analysis. Thus ancient remains have proved much more
difficult to study from the y-DNA phylogenetic point of view.
Autosomal DNA
Inside the nucleus of every cell,
each of us have 23 chromosomes. One is your sex chromosome, determining your
gender. The other 22 are your autosomal chromosomes. These contain the DNA that
codes for proteins, which are needed for growth, and for the replacement of old
worn-out cells. For an organism to grow and function properly, cells must
constantly divide to produce new cells to replace these, old, worn-out cells.
During cell division, it is essential that DNA remains intact and evenly
distributed among cells. Chromosomes are a key part of the process that ensures
DNA is accurately copied and distributed in the vast, majority, of cell
divisions. Still, mistakes do occur on rare occasions.
These mistakes or mutations are
what cause single nucleotide polymorphisms (SNPs), already discussed in the
sections above on mtDNA and y-DNA.
Autosomal DNA can be used to find
unknown relatives through commercial DNA testing, or to link modern or fossil
individuals to ancient populations.
But hang on a minute, as we each
receive 50% of our DNA from each parent, about 25% of our DNA from each of our
4 grandparents and approximately 12.5% of our DNA from each of our great
grandparents, surely this serial dilution, affects how far back you can trace
ancestry doesn’t it? Well you can test this idea: you have about 3 Billion
base pairs on your 22 chromosomes, so by generation 33 you will have, on
average, just one segment of DNA from any particular ancestor. By generation
45, that drops to 0.00017 segments.
If you think about it, an
ancestor who lived 20,000 years ago is roughly 800 generations removed from
yourself (if each generation is counted as 25 years). Therefore, through this
process of halving, the amount you receive from a particular, ancestor, will
have gone down to about 3 x 10-790 – an exceptionally small number!
Then surely tracing our autosomal
DNA to a particular, ancestor, way back in time is therefore impossible, isn’t
it?
Well yes and no! There is the
process of genetic bottlenecking to consider. When a population, for whatever
reason, is reduced to a small size, and then isolated, after a few generations
through interbreeding, all members of that population have an extremely, large
proportion of the same autosomal DNA. Imagine now, that this population is
saved from the brink of extinction and grows again, perhaps due to improved
environmental conditions, now that autosomal DNA becomes fixed within that
population.
Project that population forward
in time. The population still has the same autosomal DNA, or significant
stretches of it – some new mutations may have occurred, especially over
thousands of years.
The situation remains unchanged
until this isolated population is contacted by another and admixture of genes
occurs. A well-known example being Native Americans in the pre-Columbian
contact period, or isolated Siberian tribes up until the 19th
century.
Then along comes autosomal DNA
testing. Now we can check how many SNPs we share with many populations from
around the world, even extinct ones, whose DNA has been recovered from skeletal
remains.
Basically, the number of Alleles
(in particular SNPs) or contiguous stretches of DNA measured in centi-Morgans,
we share with a population can tell us to which populations we are related to.
Even ancient ones.
I must stress however, that this, is a simplistic explanation of how stretches of intact autosomal DNA can survive many generations. It is also, not the only, mechanism to transmit longer than expected stretches of DNA.
I must stress however, that this, is a simplistic explanation of how stretches of intact autosomal DNA can survive many generations. It is also, not the only, mechanism to transmit longer than expected stretches of DNA.
Once again, this technique is now
used to map the origins of many haplogroups through their SNPs to specific
ancient populations. Therefore, many of these groups now have simple acronyms
to show general geographic areas or indicate lifeways. A partial list is
included below:
ANE - Ancient North Eurasian
ASE - Ancient/Ancestral South
Eurasian
ASI - Ancient/Ancestral South
Indian
Austronesian – meaning
populations speaking a family of languages spoken in an area extending from
Madagascar in the west to the Pacific islands in the east.
Basal Eurasian - a hypothetical
lineage, which probably existed amount among ancient Near East individuals, who
were recent migrants out of Africa
EAS – East Asian
CHG - Caucasus Hunter Gatherers
EHG - Eastern Hunter-Gatherer
ENF - Early Neolithic Farmer, a
late Neolithic group from the near east
Khoisan - Southern Africa
Melanesian - a subregion of
Oceania/Australasia extending from the western end of the Pacific Ocea, and
eastward to Fiji.
SEA - South East Asian
SSA - Sub-Saharan African
AP - - Ancient Palaeosiberian or
just Palaeosiberian
WHG - Western Hunter-Gatherer
Then there is the autosomal DNA
from ancient individuals, whose SNP sets may show up in later populations and
thus help map ancient migrations. Again, a partial list:
Motola 12 ca. 6,000BP (Sweden)
LaB: LaBrana ca. 7,000BP (Spain)
Los: Loschbour ca. 8,000BP (Luxembourg)
Anzick1:
ca. 12,600BP (Montana USA)
AG3: Afontova Gora ca. 17,000BP
(Siberia)
MA1: the Mal'ta boy ca. 24,000BP
(Siberia)
Salkhit: ca. 34,500BP (Mongolia,
China)
GoyetQ116-1: ca. 35,000BP
(Belgium)
Kostenki 14 ca. 37,000BP
(southwest Russia)
Oase 1 ca. 39,000BP (Romania)
Tianyuan ca. 40,000BP (Beijing
China)
Ust'-Ishim ca. 45,000BP (Siberia)
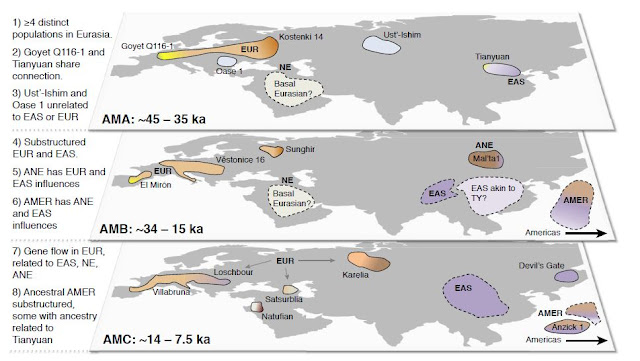
Diagram from Yang and Fu (2018)
showing the distribution of some ancient samples and groups over time.
The DNA of other, human species
have also been sequenced. In Neanderthal, Denisovans and Sima de los Huesos
hominins have had some sequences or even full genomes recovered from their
remains. Amazingly, SNPs from some of, these ancient hominins have also been
found in modern populations!
Now I feel somewhat more equipped
to read genetics papers and comment on how this evidence has been used to
indicate the timing and route(s) of the peopling of the Americas, I will
attempt something soon. Watch this space.
References
Bettinger BT, Wayne DP (2016).
Genetic Genealogy in Practice. Arlington, VA: National Genealogical Society.
Fu, Q., Posth, C., Hajdinjak, M.,
Petr, M., Mallick, S., Fernandes, D., Furtwängler, A., Haak, W., Meyer, M.,
Mittnik, A. and Nickel, B., 2016. The genetic history of ice age Europe.
Nature, 534(7606), p.200.
Wikipedia commons (2019) at: https://en.wikipedia.org/wiki/Mitochondrial_DNA
Accessed 05.03.19
Yang, M.A. and Fu, Q., 2018.
Insights into modern human prehistory using ancient genomes. Trends in
Genetics, 34(3), pp.184-196.
Bibliography:
International Society of Genetic
Genealogy at: https://isogg.org/wiki/Ancient_DNA
Genealogical DNA test at: https://en.wikipedia.org/wiki/Genealogical_DNA_test#Y_chromosome_(Y-DNA)_testing
US National Library of Medicine at: https://ghr.nlm.nih.gov/primer/basics/howmanychromosomes
Are SNPs and alleles the same thing? From Stack Exchange at:
Autosomal DNA test from UCL at: https://www.ucl.ac.uk/mace-lab/debunking/understanding-accordion/autosomal-test
Autosomal DNA, Ancient Ancestors, Ethnicity and the
Dandelion, by Roberta Estes at: https://dna-explained.com/2013/08/05/autosomal-dna-ancient-ancestors-ethnicity-and-the-dandelion/



No comments:
Post a Comment